VanOsta2024
Model summary
|
|
The VanOsta2024 model was initially introduced by van Osta et al.[1]. It is founded on the CircAdapt model published by Walmsley in 2015 Walmsley et al.[2]. On this page, a general description is given on the model. Tutorials for this model can be found here.
Physiological Background
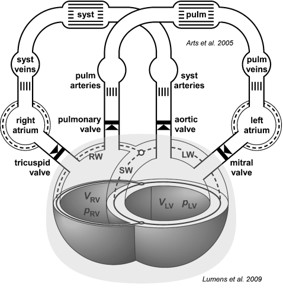
In a normal adult, the heart has four chambers, the left and right atrium and the left and right ventricle. Two valves connect the atria with the ventricles. The tricuspid valve is positioned between the right atrium and the right ventricle, and the mitral valve between the left atrium and the left ventricle. Besides these atrio-ventricular inlet valves, each ventricle has a ventricular-arterial outlet valve connecting them to the systemic and pulmonary circulation. The pulmonary valve lies between the right ventricle and the pulmonary artery, and the aortic valve lies between the left ventricle and the aorta. Wall strain determines the wall tension resulting from the passive and active material behavior of the myocardial tissue. Wall tension together with geometry determine the cavity pressure.
The pressure in a heart chamber is determined by the cavity volume. The cavity volume determines the myofibers strain. Myofiber stress arises from myofiber strain. The myofibers have a well-documented helical arrangement in the ventricles (Streeter Jr et al.[3]), and have a considerably more complex arrangement in the atria (Ho and Sanchez-Quintana[4]). Besides, the myofibers are loosely arranged into sheet-like structures. Myofiber stress is a summation of active stress, generated by the sarcomere, and passive stress resulting from stretch of elastic structures, primarily attributed to the extracellular matrix (ECM). The stress-strain relationship of the myocardial tissue is discussed in more detail in the documentation for the sarcomere module. The atria may be considered as simple cavities, encapsulated each by a relatively thin contractile elastic wall. In the CircAdapt model (Arts et al.[5], Lumens et al.[6]), each atrium is simulated by a Chamber-module. The ventricular unit is more complicated, because the right and left ventricle interact by their shared wall, the septum (Lumens et al.[6], Lumens et al.[7]). In the CircAdapt model, the composition of the left and right ventricle is simulated by a TriSegmodule. Mechanical interaction of the atrial walls with each other and with the ventricular walls probably occurs in the human heart, but is neglected because these interactions are relatively insignificant (Goldstein et al.[8], Henein et al.[9]).
In many examples of cardiac pathology, mechanical properties are inhomogeneously distributed over the various walls. To simulate such inhomogeneities in CircAdapt, the wall has been subdivided in a number of patches, each having specific mechanical properties. A patch is modeled by a Patch-module, allowing to have specific timing of activation, specific contractile properties and specific passive elastic properties. The Wall links the Patch mechanics to global hemodynamics.
Model Description
This model represents a rapid biomechanical lumped-parameter approach to simulate the intricacies of the heart and circulatory system. It uses a one-fiber model (Chamber and Wall) to establish the wall stress and cavity pressure relationship. The TriSeg module facilitates inter-ventricular interaction across the Intra-Ventricular Septum (IVS) (TriSeg). Phenomenological material laws define the active and passive stress–strain relationship in the spherical walls (Patch). The MultiPatch module introduces regional heterogeneity of tissue properties within a single wall (Wall).
Additionally, a reduced-order circulation is implemented, with systemic and pulmonary arteries and veins modeled using a single Tube0D each. The flow across systemic and pulmonary capillaries is simulated using the ArtVen module. Flow from atria to ventricles, from ventricles to arteries, and from veins to atria are simulated by means of the Valve module.
At a global scale, the Timings module manages atrial and ventricular activation. Homeostatic control can be regulated through PressureFlowControl when activated.
Default parameterization
The model is initiated with a default parameterization which represents healthy physiology. This can be visualized using the default plot statement as shown below and can be generated using the following code:
1import circadapt
2import matplotlib.pyplot as plt
3
4model = circadapt.VanOsta2024()
5model.run(stable=True)
6model.plot()
7plt.show()
(Source code, png, hires.png, pdf)

Fig. 18 Default visualization of the model with its reference parameterization.
This default parameterization gives the following scalar output:
Model |
Literature |
Unit |
Source |
|
|---|---|---|---|---|
LVEDV |
120.6 |
121 pm 31 |
mL |
Addetia et al.[10] |
LVESV |
48.3 |
47 pm 15 |
mL |
Addetia et al.[10] |
LVEF |
59.9 |
61 pm 5 |
% |
Addetia et al.[10] |
RVEDV |
108.2 |
91 [61 - 150] |
mL |
Maffessanti et al.[11] |
RVESV |
35.9 |
35 [16 - 72] |
mL |
Maffessanti et al.[11] |
RVEF |
66.8 |
62 [47 - 77] |
% |
Maffessanti et al.[11] |
SBP |
119.1 |
121 pm 12 |
mmHg |
Addetia et al.[10] |
DBP |
70.3 |
74 pm 9 |
mmHg |
Addetia et al.[10] |
mPAP |
16.1 |
[8 - 20] |
mmHg |
Humbert et al.[12] |
mLAP |
6.5 |
0 - 15 |
mmHg |
Humbert et al.[12] |
mRAP |
1.9 |
2 - 6 |
mmHg |
Humbert et al.[12] |
Model assumptions and parameterizations
Todo
Explain the solving strategy.
Present the initial parameterization.
Discuss the model verification process.
Explain adaptation protocol
Solving strategy
Todo
Update overview of the framework and its solving strategy.
Each object in this model is derived from one of the CircAdapt modules. An overview of all these objects is shown in below. All Modules together form the ordinary differential equations, which are solved using one of the implemented Solvers. By default, this set is solved using a Adams Moulton solver with a time stepsize of 0.001ms.
References
Python implementation
- class circadapt.VanOsta2024(solver: str = None, path_to_circadapt: str = None, model_state: dict = None)[source]
Bases:
Model,ModelAdapt- adapt(options: dict = {}, output: bool = False, verbose: bool = False) None
Run adaptation protocol.
The adaptation protocol runs n_cycles cycles. Each cycle has two phases, namely rest adaptation and exercise adaptation. First, in exercise, the Patches and vessel wall volumes are adapted to load. In rest, vessels are adapted to flow.
Parameters
- optionsdictionary, optional
Options for the protocol. To set options, use the function get_adapt_options(), which is used by default. The default is {}.
- outputbool, optional
DESCRIPTION. The default is False.
Returns
- senses_resultsTYPE
Only when output=True.
- senses_normTYPE
Only when output=True.
- actor_resultsTYPE
Only when output=True.
- actor_vwallTYPE
Only when output=True.
- adapt_exercise(options)
Trigger all excercise adaptation functions.
- adapt_rest(options)
Trigger all rest adaptation functions.
- add(comp_type: str) bool
- add_component(comp_type: str, comp_name: str, base: str = '') bool
Add component to CircAdapt object.
Parameters
- comp_type: str
Type of object to create in the ComponentFactory
- comp_name: str
Name of the new object to create
- base: char, optional (default=’’)
Parent object of new component
Returns
is_success (bool)
- add_smart_component(smart_component, base=None, **kwargs)
- assert_1_beat()
Test status after 1 beat, used in unit tests.
- build()[source]
Build model.
Add and link components in the model. By default, the model is empty. This function will be used in derived model builts.
- build_artven(base=None, **kwargs)
- build_heart(base=None, options=None)
- build_pfc(base=None, options=None)
- build_timings(base=None, options=None)
- calculate_and_set_matrix(verbose=False)
- check_build()
- copy()
Return a copy of itself.
- get(*arg, **kwarg)
- get_adapt_options()
Get default adapt options.
- get_matrix(verbose=False)
- get_model_reference(model_state: any = None) dict
Return reference model state as a dictionary.
If model_state is string, it is assumed to be a file location. This file will be loaded and returned. If is none, the default reference will be loaded. If no default reference available, it will be created and stored in the current active directory.
Parameters
- model_statestr or None, optional
Location of model state. The default is None.
Raises
- ValueError
DESCRIPTION.
Returns
- dict
Dictonary with model state.
- health_check()
Manually perform tests to check compatibility of model with version.
- install()
- is_stable() bool
Check if simulation is hemodynamically stable.
- load(filename: str) None
Load a model dataset from a filename.
Parameters
- filename: str
Path to file that will be loaded. The extension must be .npy or .mat.
- load_plugin_components(path_to_library: str)
Load a plugin
Parameters
- path_to_librarystr
Path to library.
- load_reference()
Load reference of current model from the package.
- model_export(style: str = None) dict
Return the stored model state.
Parameters
- style: str (optional)
Style to follow. If not given, the default style from the model is used.
Returns
dict
- model_import(model_state, check_model_state=False) None
Load model_state into CircAdapt object.
Style and model_state version is automatically recognized.
Parameters
- model_state: dict
model_state with data to set
- obj: str
Object to set
- run(*arg, **kwarg)
- save(filename: str, ext: str = None) None
Save model to filename with extention.
Parameters
- filename: str
Filename to save file to. Filename must have extention .npy or .mat
- ext: str (optional)
Extention of the file. If not given, the last 3 letters of the filename is used to determine the save method.
- set(*arg, **kwarg)
- set_component(par, *arg, **kwarg)
- trigger(par) bool
Trigger a function.
Objects in the model may have a function that can be triggered. This can be done by triggering the function using the ‘par’ parameter similar to the set function, only without a parameter.
Parameters
- parstr
Function in object that will be triggered. It has a similar form to the set/get par, i.e. ‘Component.subcomponent.function_name’
Returns
is_success (bool)
class VanOsta2024(Model, ModelAdapt):
def __init__(self,
solver: str = None,
path_to_circadapt: str = None,
model_state: dict = None,
):
if solver is None:
solver = 'adams_moulton'
self._local_save_reference = True
ModelAdapt.__init__(self)
Model.__init__(self,
solver,
path_to_circadapt=path_to_circadapt,
model_state=model_state,
)
def _update_settings(self):
# rename model components for easy input/output
self._module_rename['PressureFlowControl'] = 'PFC'
def build(self):
pass
# Circulation
self.add_smart_component('ArtVen', build='SystemicCirculation')
self.add_smart_component('ArtVen', build='PulmonaryCirculation')
self.add_smart_component('Heart', patch_type='Patch', valve_type='Valve')
self.add_smart_component('Timings')
self.add_smart_component('PressureFlowControl')
self.set_component('PFC.circulation_volume_object', 'SyArt')
self.set_component('PFC.circulation_volume_object', 'SyVen')
self.set_component('PFC.circulation_volume_object', 'PuArt')
self.set_component('PFC.circulation_volume_object', 'PuVen')
self.set_component('PFC.circulation_volume_object', 'Peri.La')
self.set_component('PFC.circulation_volume_object', 'Peri.Ra')
self.set_component('PFC.circulation_volume_object', 'Peri.TriSeg.cLv')
self.set_component('PFC.circulation_volume_object', 'Peri.TriSeg.cRv')
# # manually set papillary muscles
self.set_component("Peri.RaRv.wPapMus", "Peri.TriSeg.wRv")
self.set_component("Peri.LaLv.wPapMus", "Peri.TriSeg.wLv")
def set_reference(self):
self['Chamber']['buckling'] = True
self['Valve']['soft_closure'] = True
self['Valve']['papillary_muscles'] = False
self['Solver']['dt'] = 0.001
self['Solver']['dt_export'] = 0.002
self.set('Solver.order', 2)
self.set('Model.t_cycle', 0.85)
self['ArtVen']['p0'] = np.array([6306.25832487, 1000. ])
self['ArtVen']['q0'] = np.array([4.5e-05, 4.5e-05])
self['ArtVen']['k'] = np.array([1., 2.])
self['Tube0D']['l'] = np.array([0.4, 0.4, 0.2, 0.2])
self['Tube0D']['A_wall'] = np.array([1.12362733e-04, 6.57883944e-05, 9.45910889e-05, 8.22655361e-05])
self['Tube0D']['k'] = np.array([1.66666667, 2.33333333, 1.66666667, 2.33333333])
self['Tube0D']['p0'] = np.array([12162.50457811, 287.7083132 , 2132.51755623, 830.54673184])
self['Tube0D']['A0'] = np.array([0.0004983 , 0.00049909, 0.00047138, 0.00050803])
self['Tube0D']['target_wall_stress'] = np.array([500000., 500000., 500000., 500000.])
self['Tube0D']['target_mean_flow'] = np.array([0.17, 0.17, 0.17, 0.17])
self['Bag']['k'] = np.array([10.])
self['Bag']['p_ref'] = np.array([1000.])
self['Bag']['V_ref'] = np.array([0.00054267])
self['Patch']['Am_ref'] = np.array([0.00425687, 0.00401573, 0.00966859, 0.00289936, 0.01084227])
self['Patch']['V_wall'] = np.array([4.46069398e-06, 2.14548521e-06, 7.35720515e-05, 1.88904978e-05, 3.67720116e-05,])
self['Patch']['v_max'] = np.array([14., 14., 7., 7., 7.])
self['Patch']['l_se0'] = np.array([0.04, 0.04, 0.04, 0.04, 0.04])
self['Patch']['l_s0'] = np.array([1.8, 1.8, 1.8, 1.8, 1.8])
self['Patch']['l_s_ref'] = np.array([2., 2., 2., 2., 2.])
self['Patch']['dl_s_pas'] = np.array([0.6, 0.6, 0.6, 0.6, 0.6])
self['Patch']['Sf_pas'] = np.array([2248.53598 , 2684.76100348, 731.24545453, 729.06063771, 749.47694522,])
self['Patch']['tr'] = np.array([0.4 , 0.4 , 0.25, 0.25, 0.25])
self['Patch']['td'] = np.array([0.4 , 0.4 , 0.25, 0.25, 0.25])
self['Patch']['time_act'] = np.array([0.15 , 0.15 , 0.425, 0.425, 0.425])
self['Patch']['Sf_act'] = np.array([ 84000., 84000., 120000., 120000., 120000.])
self['Patch']['fac_Sf_tit'] = np.array([0.01, 0.01, 0.01, 0.01, 0.01])
self['Patch']['k1'] = np.array([10., 10., 10., 10., 10.])
self['Patch']['dt'] = np.array([0., 0., 0., 0., 0.])
self['Patch']['C_rest'] = np.array([0., 0., 0., 0., 0.])
self['Patch']['l_si0'] = np.array([1.51, 1.51, 1.51, 1.51, 1.51])
self['Patch']['LDAD'] = np.array([1.057, 1.057, 1.057, 1.057, 1.057])
self['Patch']['ADO'] = np.array([0.65, 0.65, 0.65, 0.65, 0.65])
self['Patch']['LDCC'] = np.array([4., 4., 4., 4., 4.])
self['Patch']['SfPasMaxT'] = np.array([320000., 320000., 16000., 16000., 16000.])
self['Patch']['SfPasActT'] = np.array([40000., 40000., 20000., 20000., 20000.])
self['Patch']['FacSfActT'] = np.array([0.8, 0.8, 1. , 1. , 1. ])
self['Patch']['LsPasActT'] = np.array([2.42, 2.42, 2.42, 2.42, 2.42])
self['Patch']['adapt_gamma'] = np.array([0.5, 0.5, 0.5, 0.5, 0.5])
self['Patch']['transmat00'] = np.array([-0.5751, -0.5751, -0.5751, -0.5751, -0.5751])
self['Patch']['transmat01'] = np.array([-0.7851, -0.7851, -0.7851, -0.7851, -0.7851])
self['Patch']['transmat02'] = np.array([0.6063, 0.6063, 0.6063, 0.6063, 0.6063])
self['Patch']['transmat03'] = np.array([-0.5565, -0.5565, -0.5565, -0.5565, -0.5565])
self['Patch']['transmat10'] = np.array([-0.1279, -0.1279, -0.1279, -0.1279, -0.1279])
self['Patch']['transmat11'] = np.array([0.0999, 0.0999, 0.0999, 0.0999, 0.0999])
self['Patch']['transmat12'] = np.array([0.2066, 0.2066, 0.2066, 0.2066, 0.2066])
self['Patch']['transmat13'] = np.array([-1.8441, -1.8441, -1.8441, -1.8441, -1.8441])
self['Patch']['transmat20'] = np.array([-0.1865, -0.1865, -0.1865, -0.1865, -0.1865])
self['Patch']['transmat21'] = np.array([-0.201, -0.201, -0.201, -0.201, -0.201])
self['Patch']['transmat22'] = np.array([1.3195, 1.3195, 1.3195, 1.3195, 1.3195])
self['Patch']['transmat23'] = np.array([-11.8745, -11.8745, -11.8745, -11.8745, -11.8745])
self['Valve']['adaptation_A_open_fac'] = np.array([1., 1., 1., 1., 1., 1.])
self['Valve']['A_open'] = np.array([0.00049916, 0.00047155, 0.00047155, 0.00050805, 0.00049835,
0.00049835])
self['Valve']['A_leak'] = np.array([0.00049916, 1.e-09, 1.e-09, 0.00050805, 1.e-09, 1.e-09])
self['Valve']['l'] = np.array([0.01260512, 0.01225144, 0.01225144, 0.01271679, 0.01259479,
0.01259479])
self['Valve']['L_fac_prox'] = np.array([0.75, 0.75, 0.75, 0.75, 0.75, 0.75])
self['Valve']['L_fac_dist'] = np.array([0.75, 0.75, 0.75, 0.75, 0.75, 0.75])
self['Valve']['L_fac_valve'] = np.array([1.5, 1.5, 1.5, 1.5, 1.5, 1.5])
self['Valve']['rho_b'] = np.array([1050., 1050., 1050., 1050., 1050., 1050.])
self['Valve']['papillary_muscles'] = np.array([False, False, False, False, False, False])
self['Valve']['papillary_muscles_slope'] = np.array([100., 100., 100., 100., 100., 100.])
self['Valve']['papillary_muscles_min'] = np.array([0.1, 0.1, 0.1, 0.1, 0.1, 0.1])
self['Valve']['papillary_muscles_A_open_fac'] = np.array([0.1, 0.1, 0.1, 0.1, 0.1, 0.1])
self['Valve']['soft_closure'] = np.array([ True, True, True, True, True, True])
self['Valve']['fraction_A_open_Aext'] = np.array([0.9, 0.9, 0.9, 0.9, 0.9, 0.9])
self['Timings']['time_fac'] = np.array([1.])
self['Timings']['tau_av'] = np.array([0.150025])
self['Timings']['dtau_av'] = np.array([0.])
self['Timings']['law_tau_av'] = np.array([1])
self['Timings']['law_Ra2La'] = np.array([1])
self['Timings']['law_ta'] = np.array([1])
self['Timings']['law_tv'] = np.array([1])
self['Timings']['c_tau_av0'] = np.array([0.])
self['Timings']['c_tau_av1'] = np.array([0.1765])
self['Timings']['c_ta_rest'] = np.array([0.])
self['Timings']['c_ta_tcycle'] = np.array([0.17647059])
self['Timings']['c_tv_rest'] = np.array([0.1])
self['Timings']['c_tv_tcycle'] = np.array([0.4])
self['PFC']['p0'] = np.array([12200.])
self['PFC']['q0'] = np.array([8.5e-05])
self['PFC']['stable_threshold'] = np.array([0.0001])
self['PFC']['is_active'] = np.array([ True])
self['PFC']['fac'] = np.array([1.])
self['PFC']['fac_pfc'] = np.array([1.])
self['PFC']['epsilon'] = np.array([0.4])
self['TriSeg']['tau'] = np.array([2.])
self['TriSeg']['ratio_septal_LV_Am'] = np.array([0.3])
self['TriSeg']['max_number_of_iterations'] = np.array([100.])
self['TriSeg']['thresh_F'] = np.array([0.001])
self['TriSeg']['thresh_dV'] = np.array([1.e-09])
self['TriSeg']['thresh_dY'] = np.array([1.e-06])
self.run(stable=True)
# self.adapt()
# set target volume
self['PFC']['target_volume'] = self['PFC']['circulation_volume']
return
def get_unittest_targets(self):
"""Hardcoded results after initializing and running 1 beat."""
return {
'LVEDV': 120.5,
'LVESV': 48.3,
}
def get_unittest_results(self, model):
"""Real-time results after initializing and running 1 beat."""
LVEDV = np.max(model['Cavity']['V'][:, 'cLv'])*1e6
LVESV = np.min(model['Cavity']['V'][:, 'cLv'])*1e6
return {
'LVEDV': LVEDV,
'LVESV': LVESV,
}
def plot(self, fig=None):
# TODO
self.plot_extended(fig)
def plot_extended(self, fig=None):
if len(self['Solver']['t'])==0:
raise RuntimeError('Can not plot the model, as no data is available. Did you simulate the model?')
if fig is None:
fig = 1
if isinstance(fig, int):
fig = plt.figure(fig, clear=True, figsize=(12, 8))
# Settings
grid_size = [32, 32]
def get_lim(module, signal, locs=slice(None, None, None)):
signal = self[module][signal][:, locs]
lim = np.array([np.min(signal), np.max(signal)])
lim += np.array([-1, 1]) * 0.1*np.diff(lim)
return lim
lim_V = get_lim('Cavity', 'V', ['cRv', 'Ra', 'La', 'cLv']) * 1e6
lim_V[0] = 0
lim_p = get_lim('Cavity', 'p', ['cRv', 'Ra', 'La', 'cLv']) / 133
lim_p[0] = np.min([lim_p[0], 0])
lim_Ls = get_lim('Patch', 'l_s')
lim_Sf = get_lim('Patch', 'Sf') * 1e-3
lim_q = get_lim('Valve', 'q',
['LaLv', 'RaRv', 'LvSyArt', 'RaPuArt']) * 1e6
all_lim = [lim_V, lim_p, lim_Ls, lim_Sf, lim_q]
if (np.any(np.isnan(all_lim)) or np.any(np.isinf(all_lim))):
lim_V = [0, 200]
lim_p = [0, 150]
lim_Ls = [1.5, 2.0]
lim_Sf = [0, 100]
lim_q = [-1e-3, 1e-3]
# Pressure Volume plot
axPV = plt.subplot2grid(grid_size, (0, 17), rowspan=15, colspan=15, fig=fig)
axPV.plot(self['Cavity']['V'][:, 'cLv']*1e6,
self['Cavity']['p'][:, 'cLv']/133,
c=colors['Lv'],
)
axPV.plot(self['Cavity']['V'][:, 'cRv']*1e6,
self['Cavity']['p'][:, 'cRv']/133,
c=colors['Rv'],
)
axPV.plot(self['Cavity']['V'][:, 'La']*1e6,
self['Cavity']['p'][:, 'La']/133,
c=colors['La'],
)
axPV.plot(self['Cavity']['V'][:, 'Ra']*1e6,
self['Cavity']['p'][:, 'Ra']/133,
c=colors['Ra'],
)
axPV.spines[['top', 'right']].set_visible(False)
axPV.set_title('Pressure-Volume loop', weight='bold')
axPV.set_xlabel('Volume [mL]')
axPV.set_ylabel('Pressure [mmHg]')
axPV.spines[['bottom', 'left']].set_position(('outward', 5))
ylabel_x_left = -0.25
ylabel_x_right = 1.25
# Volumes
t = self['Solver']['t']*1e3
axVRv = plt.subplot2grid(grid_size, (0, 0), rowspan=8, colspan=6, fig=fig)
axVRv.plot(t, self['Cavity']['V'][:, 'cRv']*1e6,
c=colors['Rv'],
)
axVRv.plot(t, self['Cavity']['V'][:, 'Ra']*1e6,
c=colors['Ra'],
)
axVRv.set_ylim(lim_V)
axVRv.set_ylabel('Volume\n[mL]')
axVRv.spines[['top', 'right']].set_visible(False)
axVRv.set_title('Right Heart', weight='bold')
# axVRv.set_xticks([])
axVRv.tick_params(axis='both', direction='in')
axVRv.yaxis.set_label_coords(ylabel_x_left, 0.5)
axVLv = plt.subplot2grid(grid_size, (0, 6), rowspan=8, colspan=6, fig=fig)
axVLv.plot(t, self['Cavity']['V'][:, 'cLv']*1e6,
c=colors['Lv'],
)
axVLv.plot(t, self['Cavity']['V'][:, 'La']*1e6,
c=colors['La'],
)
axVLv.set_ylabel('Volume\n[mL]')
axVLv.set_ylim(lim_V)
axVLv.yaxis.set_ticks_position('right')
axVLv.yaxis.set_label_position('right')
axVLv.spines['right'].set_position(('outward', 0))
axVLv.spines[['top', 'left']].set_visible(False)
axVLv.set_title('Left Heart', weight='bold')
# axVLv.set_xticks([])
axVLv.tick_params(axis='both', direction='in')
axVLv.yaxis.set_label_coords(ylabel_x_right, 0.5)
# Pressures
axpRv = plt.subplot2grid(
grid_size, (8, 0), rowspan=8, colspan=6, fig=fig, sharex=axVRv)
axpRv.plot(t, self['Cavity']['p'][:, 'cRv']/133,
c=colors['Rv'],
)
axpRv.plot(t, self['Cavity']['p'][:, 'Ra']/133,
c=colors['Ra'],
)
axpRv.plot(t, self['Cavity']['p'][:, 'PuArt']/133,
c=colors['PuArt'],
)
axpRv.spines[['top', 'right']].set_visible(False)
# axpRv.set_xticks([])
axpRv.tick_params(axis='both', direction='in')
axpRv.set_ylim(lim_p)
axpRv.set_ylabel('Pressure\n[mmHg]')
axpRv.yaxis.set_label_coords(ylabel_x_left, 0.5)
axpLv = plt.subplot2grid(grid_size, (8, 6), rowspan=8, colspan=6, fig=fig, sharex=axVLv)
axpLv.plot(t, self['Cavity']['p'][:, 'cLv']/133,
c=colors['Lv'],
)
axpLv.plot(t, self['Cavity']['p'][:, 'La']/133,
c=colors['La'],
)
axpLv.plot(t, self['Cavity']['p'][:, 'SyArt']/133,
c=colors['SyArt'],
)
axpLv.yaxis.set_ticks_position('right')
axpLv.yaxis.set_label_position('right')
axpLv.spines['right'].set_position(('outward', 0))
axpLv.spines[['top', 'left']].set_visible(False)
# axpLv.set_xticks([])
axpLv.tick_params(axis='both', direction='in')
axpLv.set_ylim(lim_p)
axpLv.set_ylabel('Pressure\n[mmHg]')
axpLv.yaxis.set_label_coords(ylabel_x_right, 0.5)
# Valves
ax = plt.subplot2grid(grid_size, (16, 0), rowspan=6, colspan=6, fig=fig, sharex=axVRv)
ax.plot(t, self['Valve']['q'][:, 'RaRv']*1e6,
c=colors['RaRv'],
)
ax.plot(t, self['Valve']['q'][:, 'RvPuArt']*1e6,
c=colors['RvPuArt'],
)
ax.spines[['top', 'right']].set_visible(False)
ax.set_ylim(lim_q)
# ax.set_xticks([])
ax.set_ylabel('Flow\n[mL/s]')
ax.yaxis.set_label_coords(ylabel_x_left, 0.5)
ax = plt.subplot2grid(grid_size, (16, 6), rowspan=6, colspan=6, fig=fig, sharex=axVLv)
ax.plot(t, self['Valve']['q'][:, 'LaLv']*1e6,
c=colors['LaLv'],
)
ax.plot(t, self['Valve']['q'][:, 'LvSyArt']*1e6,
c=colors['LvSyArt'],
)
ax.spines[['top', 'left']].set_visible(False)
ax.set_ylim(lim_q)
ax.yaxis.set_ticks_position('right')
ax.yaxis.set_label_position('right')
# ax.set_xticks([])
ax.set_ylabel('Flow\n[mL/s]')
ax.yaxis.set_label_coords(ylabel_x_right, 0.5)
# Stress
ax = plt.subplot2grid(grid_size, (22, 0), rowspan=4, colspan=6, fig=fig, sharex=axVRv)
ax.plot(t, self['Patch']['Sf'][:, 'pRv0']*1e-3,
c=colors['Rv'],
)
ax.plot(t, self['Patch']['Sf'][:, 'pRa0']*1e-3,
c=colors['Ra'],
)
ax.spines[['top', 'right']].set_visible(False)
# ax.set_xticks([])
ax.set_ylim(lim_Sf)
ax.set_ylabel('Total\nstress [kPa]')
ax.yaxis.set_label_coords(ylabel_x_left, 0.5)
ax = plt.subplot2grid(grid_size, (22, 6), rowspan=4, colspan=6, fig=fig, sharex=axVLv)
ax.plot(t, self['Patch']['Sf'][:, 'pLv0']*1e-3,
c=colors['Lv'],
)
ax.plot(t, self['Patch']['Sf'][:, 'pSv0']*1e-3,
c=colors['Sv'],
)
ax.plot(t, self['Patch']['Sf'][:, 'pLa0']*1e-3,
c=colors['La'],
)
ax.spines[['top', 'left']].set_visible(False)
ax.yaxis.set_ticks_position('right')
ax.yaxis.set_label_position('right')
# ax.set_xticks([])
ax.set_ylim(lim_Sf)
ax.set_ylabel('Total\nstress [kPa]')
ax.yaxis.set_label_coords(ylabel_x_right, 0.5)
# Sarcomere Length
ax = plt.subplot2grid(grid_size, (26, 0), rowspan=6, colspan=6, fig=fig, sharex=axVRv)
ax.plot(t, self['Patch']['l_s'][:, 'pRv0'],
c=colors['Rv'],
)
ax.plot(t, self['Patch']['l_s'][:, 'pRa0'],
c=colors['Ra'],
)
ax.spines[['top', 'right']].set_visible(False)
ax.spines['bottom'].set_position(('outward', 5))
ax.set_ylim(lim_Ls)
ax.set_ylabel('Sarcomere\nlength [$\\mu$m]')
ax.yaxis.set_label_coords(ylabel_x_left, 0.5)
ax = plt.subplot2grid(grid_size, (26, 6), rowspan=6, colspan=6, fig=fig, sharex=axVLv)
ax.plot(t, self['Patch']['l_s'][:, 'pLv0'],
c=colors['Lv'],
)
ax.plot(t, self['Patch']['l_s'][:, 'pSv0'],
c=colors['Sv'],
)
ax.plot(t, self['Patch']['l_s'][:, 'pLa0'],
c=colors['La'],
)
ax.spines[['top', 'left']].set_visible(False)
ax.spines['bottom'].set_position(('outward', 5))
ax.yaxis.set_ticks_position('right')
ax.yaxis.set_label_position('right')
ax.set_ylim(lim_Ls)
ax.set_ylabel('Sarcomere\nlength [$\\mu$m]')
ax.yaxis.set_label_coords(ylabel_x_right, 0.5)
# ax.set_xlabel('Time [ms]')
# ax.xaxis.set_label_coords(0, -0.3)
# Plot TriSeg
titles = ['Pre-A', 'Onset QRS', 'Peak LV \n pressure', 'AV close']
idx = [0,
np.argmax(np.diff(self['Patch']['C'][:, 'pLv0'])>0),
np.argmax(self['Cavity']['p'][:, 'cLv']),
len(t) - 1 - np.argmax(
np.diff(self['Valve']['q'][:, 'LvSyArt'][::-1])>0)
]
for i in range(4):
ax = plt.subplot2grid(grid_size, (26, 16+4*i),
rowspan=5, colspan=4, fig=fig)
triseg2022(self, ax, idx[i],
colors=[colors['Lv'], colors['Sv'], colors['Rv']])
ax.spines[['top', 'right', 'bottom', 'left']].set_visible(False)
ax.set_xticks([])
ax.set_yticks([])
plt.xlabel(titles[i], fontsize=12)
# Plot settings
plt.subplots_adjust(
top=0.96,
bottom=0.05,
left=0.075,
right=0.98,
hspace=5,
wspace=0.5)
plt.draw()
# Stress strain
ax_right_stress_strain = plt.subplot2grid(grid_size, (18, 17),
rowspan=7, colspan=7, fig=fig)
ax_left_stress_strain = plt.subplot2grid(grid_size, (18, 24),
rowspan=7, colspan=7, fig=fig)
ax_right_stress_strain.plot(
self['Patch']['Ef'][:, 'pRa0'],
self['Patch']['Sf'][:, 'pRa0']*1e-3,
c=colors['Ra'],
)
ax_right_stress_strain.plot(
self['Patch']['Ef'][:, 'pRv0'],
self['Patch']['Sf'][:, 'pRv0']*1e-3,
c=colors['Rv'],
)
ax_left_stress_strain.plot(
self['Patch']['Ef'][:, 'pLa0'],
self['Patch']['Sf'][:, 'pLa0']*1e-3,
c=colors['La'],
)
ax_left_stress_strain.plot(
self['Patch']['Ef'][:, 'pLv0'],
self['Patch']['Sf'][:, 'pLv0']*1e-3,
c=colors['Lv'],
)
ax_left_stress_strain.plot(
self['Patch']['Ef'][:, 'pSv0'],
self['Patch']['Sf'][:, 'pSv0']*1e-3,
c=colors['Sv'],
)
ylim = [np.min(self['Patch']['Sf'])*1e-3,
np.max(self['Patch']['Sf'])*1e-3]
ylim_ptp = np.ptp(ylim)*0.025
ylim[0] -= ylim_ptp
ylim[1] += ylim_ptp
ax_right_stress_strain.set_ylim(ylim)
ax_left_stress_strain.set_ylim(ylim)
xlim = [np.min(self['Patch']['Ef']),
np.max(self['Patch']['Ef'])]
xlim_ptp = np.ptp(xlim)*0.025
xlim[0] -= xlim_ptp
xlim[1] += xlim_ptp
ax_right_stress_strain.set_xlim(xlim)
ax_left_stress_strain.set_xlim(xlim)
ax_right_stress_strain.spines[['left', 'bottom']].set_position(('outward', 5))
ax_left_stress_strain.spines[['right', 'bottom']].set_position(('outward', 5))
ax_right_stress_strain.spines[['top', 'right']].set_visible(False)
ax_right_stress_strain.set_ylabel('[kPa]', rotation=0, va='bottom', ha='right')
ax_right_stress_strain.yaxis.set_label_coords(0., 1.05)
ax_right_stress_strain.set_xlabel('Strain [-]')
ax_left_stress_strain.spines[['top', 'left']].set_visible(False)
ax_left_stress_strain.yaxis.set_ticks_position('right')
ax_left_stress_strain.yaxis.set_label_position('right')
ax_left_stress_strain.set_ylabel('Stress\n[kPa]', rotation=0, va='bottom', ha='left')
ax_left_stress_strain.yaxis.set_label_coords(1., 1.05)
ax_left_stress_strain.set_xlabel('Strain [-]')