VanOsta2022 Regional Myocardial (Dys)function
The multipatch implementation of the wall module allows for simulationg regional wall mechanics. To initiate this, the wall can be split in multiple patches using the lines:
1model['Wall']['n_patch'][2:4] = [12, 6]
2
In this example, we apply a small heterogeneity in activation delay.
1model['Patch']['dt'][2:14] = np.linspace(0, 0.02, 12)
2model['Patch']['dt'][14:20] = np.linspace(0.01, 0.05, 6)
3
Running this simulation results in the following pressures, volumes, and regional fiber strain.
(Source code, png, hires.png, pdf)
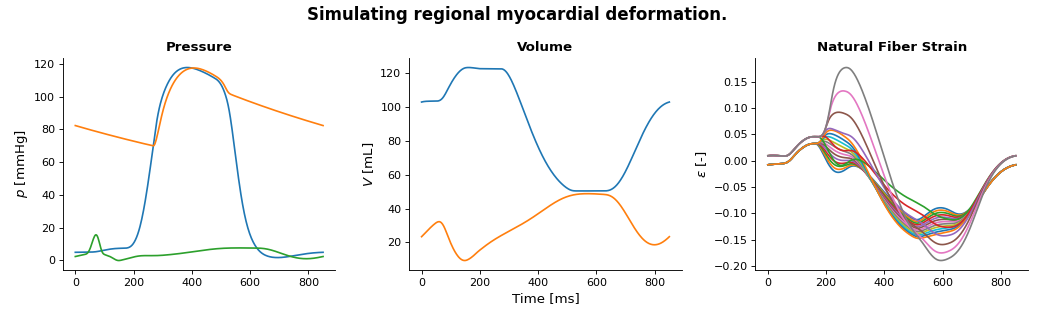
The full code to generate this plot is shown below.
1"""
2Tutorial CircAdapt VanOsta2022.
3
4March 2023, by Nick van Osta
5
6This tutorial demonstrates how to model regional mechanics, how to change
7regional parameters and how to obtain and plot regional
8"""
9
10import numpy as np
11import matplotlib.pyplot as plt
12import circadapt
13from circadapt.model import VanOsta2023
14
15# %% 1. load model
16model = VanOsta2023()
17
18# Split the LV in 12 and SV in 6 segments
19model['Wall']['n_patch'][2:4] = [12, 6]
20
21# Set activation delay
22model['Patch']['dt'][2:14] = np.linspace(0, 0.02, 12)
23model['Patch']['dt'][14:20] = np.linspace(0.01, 0.05, 6)
24
25# Run beats
26model.run(stable=True)
27
28# Plot data
29fig = plt.figure(2, figsize=(13, 4))
30ax1 = fig.add_subplot(1, 3, 1)
31ax2 = fig.add_subplot(1, 3, 2)
32ax3 = fig.add_subplot(1, 3, 3)
33
34# Plot pressure
35ax1.plot(model['Solver']['t']*1e3,
36 model['Cavity']['p'][:, ['cLv', 'SyArt', 'La']]*7.5e-3,
37 )
38
39# Plot Volume
40ax2.plot(model['Solver']['t']*1e3,
41 model['Cavity']['V'][:, ['cLv', 'La']]*1e6,
42 )
43
44# Plot natural fiber strain
45ax3.plot(model['Solver']['t']*1e3,
46 model['Patch']['Ef'][:, 2:20],
47 )
48
49# plot design, add labels
50for ax in [ax1, ax2, ax3]:
51 ax.spines[['right', 'top']].set_visible(False)
52ax2.set_xlabel('Time [ms]', fontsize=12)
53
54ax1.set_ylabel('$p$ [mmHg]', fontsize=12)
55ax2.set_ylabel('$V$ [mL]', fontsize=12)
56ax3.set_ylabel('$\epsilon$ [-]', fontsize=12)
57
58ax1.set_title('Pressure',
59 fontsize=12, fontweight='bold')
60ax2.set_title('Volume',
61 fontsize=12, fontweight='bold')
62ax3.set_title('Natural Fiber Strain',
63 fontsize=12, fontweight='bold')
64
65fig.suptitle('Simulating regional myocardial deformation. ',
66 fontsize=15, fontweight='bold')
67
68plt.tight_layout()
69plt.draw()