1"""
2Tutorial CircAdapt VanOsta2022.
3
4November 2022, by Nick van Osta
5
6The goal of this tutorial is to understand the CircAdapt framework and to use
7the VanOsta2022 model. This tutorial assumes little to no knowledge about
8python. Therefore, basic python conventions and syntax will be discussed.
9This tutorial assumes the installation is followed as described on the wiki
10(https://wiki.circadapt.org/index.php?title=Circadapt_in_Python). This uses
11Python >3.9 installed with anaconda and editted in Spyder. Other ways are
12possible, but might not be in line with this tutorial.
13
14Content
15-------
16 1. Basics of python
17 2. Load the model
18 3. Plot global hemodynamics
19 4. Change parameters
20 5. Multipatch and local dynamics
21 6. Save and Load
22"""
23
24# Uncomment next lines if not installed
25# import sys
26# sys.path.append('../../../src/')
27
28import circadapt
29
30# Uncomment next lines if not installed
31# circadapt.DEFAULT_PATH_TO_CIRCADAPT = "../../../CircAdapt_Library/out/build/x64-Release/CircAdaptLib.dll"
32
33# %% 1. Basics of python
34print('1. Basics of python')
35
36# Always start the document with importing modules.
37# Numpy is used for mathematics, Matplotlib for plots. These are conventionally
38# imported as np and plt.
39import numpy as np
40import matplotlib.pyplot as plt
41
42# Only import the class we need in this tutorial.
43from circadapt.model import VanOsta2023
44# CA = VanOsta2022()
45
46# alternatively, you can import the whole package, but it changes the way you
47# make the object.
48# import circadapt
49# CA = circadapt.VanOsta2022()
50
51# Parameters types are automaticaly set or changed by the interpreter, but it
52# is good to create an integer when you need an integer and float when needed.
53i = 1 # integer
54f = 1. # float
55b = True # bool
56l = [1, 2, 3] # list
57d = {'a': 1, 'b': 2} # dictionary
58
59# get data from the list and dictionary
60first_item_of_list = l[0]
61item_from_dictionary = d['a']
62
63# the use of numpy is advised for more complex use and for calculation
64numpy_array = np.array(l)
65print('Find if array is 2: ', (numpy_array == 2))
66print('Multiply array with 2: ', (numpy_array * 2))
67
68# In spyder, you can place bullits. While debugging, ipython will stop at these
69# bullits. You can also press f9 to run a single line or selection and press
70# crtl+<enter> to run a block seperated by # %%
71
72# %% 2. load model
73print('\n 2. Load model. ')
74# Load predefined model with predefined parameterization
75# More information on this model can be found here:
76# https://wiki.circadapt.org/index.php?title=VanOsta2022
77CA = VanOsta2023()
78
79# CircAdapt tries to follow the syntax of python and numpy as much as possible.
80# The object can be handled as a dictionary. Content can be printed in the
81# console, and printed on request in the ipython console.
82print('The result of printing the object gives information about the '
83 'components: ')
84print(CA)
85
86# components can also be retrieved as a list of strings
87components = CA.components
88print('Components of this model: ', components, '\n')
89
90# Similar to the object itself, components can be printed
91print('Patches in this model: ')
92print(CA['Patch2022'])
93
94# Each component points to multiple c++ objects of that component type. The
95# objects can also be obtained using
96objects = CA['Patch2022'].objects
97parameters = CA['Patch2022'].parameters
98signals = CA['Patch2022'].signals
99
100# Signals are not stored, so they are only available after running a beat.
101# Therefore the model should run. You can either run a number of beats, or run
102# until the model is hemodynamically stable.
103CA.run(5)
104CA.run(stable=True)
105
106# %% 3. Plot global hemodynamics
107# Here is an example code to plot the PV loop
108# First we open a figure. Assigning this figure to a variable is optional, but
109# is useful for design purposes.
110fig = plt.figure(1)
111
112# get volume and pressure of LV
113Vlv = CA['Cavity']['V'][:, 'cLv']*1e6
114plv = CA['Cavity']['p'][:, 'cLv']*7.5e-3
115
116# get volume and pressure of RV
117Vrv = CA['Cavity']['V'][:, 'cRv']*1e6
118prv = CA['Cavity']['p'][:, 'cRv']*7.5e-3
119
120# You can also use location names to get/set signals and parameters
121# For this, use only the last part of the full object name, e.g. cLv for
122# Model.Peri.TriSeg.cLv. You can get one signal or multiple signals
123Vlv = CA['Cavity']['V'][:, 'cLv']*1e6
124Vrv = CA['Cavity']['V'][:, 'cRv']*1e6
125pressure = CA['Cavity']['p'][:, ['cLv', 'cRv']]*7.5e-3
126
127# you can split the two pressure signals into two parameters using the
128# following line. First transpose the pressure such that the first axis sets
129# the signals
130plv, prv = pressure.T
131
132# Now we plot the two lines.
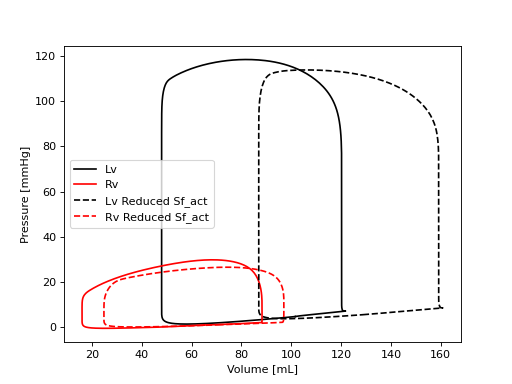
133line1 = plt.plot(Vlv, plv, c='k', label='Lv')
134line2 = plt.plot(Vrv, prv, c='r', label='Rv')
135plt.ylabel('Pressure [mmHg]')
136plt.xlabel('Volume [mL]')
137plt.legend()
138
139# %% 4. Change parameters
140# Now reduce the contractility of all 3 ventricular walls, run the simulation,
141# and plot the data
142CA['Patch2022']['Sf_act'][2:] = 60e3
143CA.run(stable=True)
144plt.plot(CA['Cavity']['V'][:, 'cLv']*1e6, CA['Cavity']['p'][:, 'cLv']*7.5e-3, 'k--', label='Lv Reduced Sf_act')
145plt.plot(CA['Cavity']['V'][:, 'cRv']*1e6, CA['Cavity']['p'][:, 'cRv']*7.5e-3, 'r--', label='Rv Reduced Sf_act')
146plt.legend()
147
148
149# %% 5. Multipatch and local dynamics
150# Set up a new multipatch model and set an activation delay
151CA_multipatch = VanOsta2023()
152
153# The number of patches is specified in the wall. Here, we set 12 Lv patches
154# and 6 Sv patches. Then, we change the dt in these patches.
155CA_multipatch['Wall2022']['n_patch'][2:4] = [12, 6]
156CA_multipatch['Patch2022']['dt'][2:14] = np.linspace(0, 0.01, 12)
157CA_multipatch['Patch2022']['dt'][14:20] = np.linspace(0, 0.01, 6)
158
159# Run beats
160CA_multipatch.run(stable=True)
161
162# Plot data
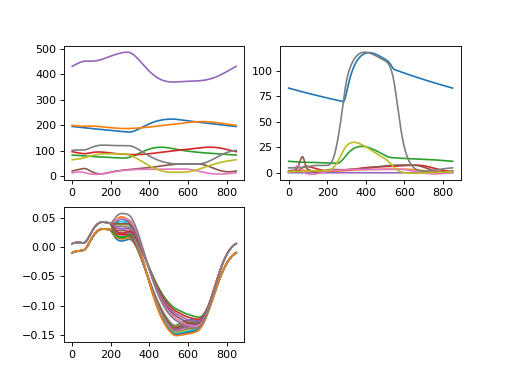
163fig = plt.figure(2)
164
165# In the first subplot, plot all volumes
166ax1 = plt.subplot(2, 2, 1)
167plt.plot(CA_multipatch['Solver']['t']*1e3,
168 CA_multipatch['Cavity']['V']*1e6,
169 )
170# in the second subplot, plot all pressures
171ax1 = plt.subplot(2, 2, 2)
172plt.plot(CA_multipatch['Solver']['t']*1e3,
173 CA_multipatch['Cavity']['p']*7.5e-3,
174 )
175# in the third subplot, plot all natural fiber strains.
176ax1 = plt.subplot(2, 2, 3)
177plt.plot(CA_multipatch['Solver']['t']*1e3,
178 CA_multipatch['Patch2022']['Ef'][:, 2:20],
179 )
180
181# %% 6. Save and Load
182# Simulations can be saved and loaded using the following code.
183CA_reference = VanOsta2023()
184
185# use .npy extension in filename
186CA_reference.save('reference.npy')
187
188# ander bestand
189model = VanOsta2023()
190model.load('reference.npy')
191
192# if you want to save and load a structure without writing it to a file, use
193# the follow lines. Note that signals are not filled, you have to run at least
194# 1 beat.
195data = CA_reference.model_export()
196CA_reference.model_import(data)